SimpleITK Images, They're Physical Objects 
SimpleITK conventions:
- Image access is in x,y,z order, image.GetPixel(x,y,z) or image[x,y,z], with zero based indexing.
- If the output of an ITK filter has non-zero starting index, then the index will be set to 0, and the origin adjusted accordingly.
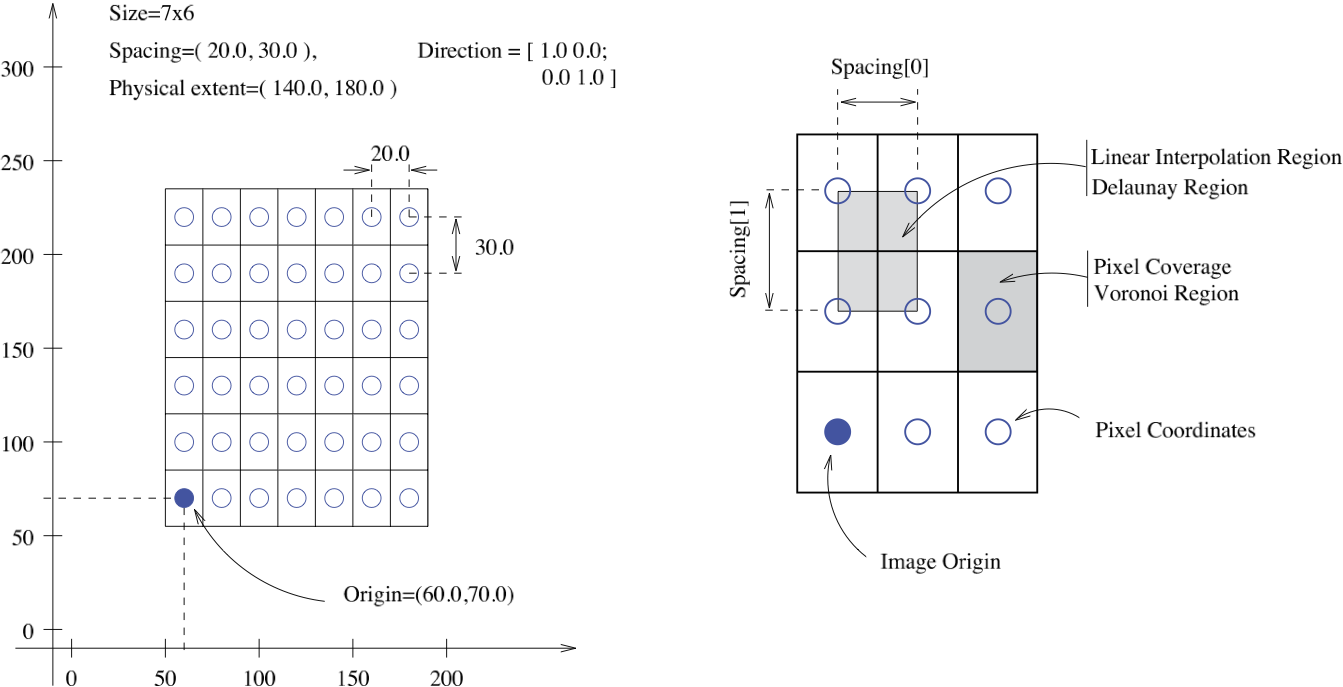
The unique feature of SimpleITK (derived from ITK) as a toolkit for image manipulation and analysis is that it views images as physical objects occupying a bounded region in physical space. In addition images can have different spacing between pixels along each axis, and the axes are not necessarily orthogonal. The following figure illustrates these concepts.
Pixel Types¶
The pixel type is represented as an enumerated type. The following is a table of the enumerated list.
| sitkUInt8 | Unsigned 8 bit integer |
| sitkInt8 | Signed 8 bit integer |
| sitkUInt16 | Unsigned 16 bit integer |
| sitkInt16 | Signed 16 bit integer |
| sitkUInt32 | Unsigned 32 bit integer |
| sitkInt32 | Signed 32 bit integer |
| sitkUInt64 | Unsigned 64 bit integer |
| sitkInt64 | Signed 64 bit integer |
| sitkFloat32 | 32 bit float |
| sitkFloat64 | 64 bit float |
| sitkComplexFloat32 | complex number of 32 bit float |
| sitkComplexFloat64 | complex number of 64 bit float |
| sitkVectorUInt8 | Multi-component of unsigned 8 bit integer |
| sitkVectorInt8 | Multi-component of signed 8 bit integer |
| sitkVectorUInt16 | Multi-component of unsigned 16 bit integer |
| sitkVectorInt16 | Multi-component of signed 16 bit integer |
| sitkVectorUInt32 | Multi-component of unsigned 32 bit integer |
| sitkVectorInt32 | Multi-component of signed 32 bit integer |
| sitkVectorUInt64 | Multi-component of unsigned 64 bit integer |
| sitkVectorInt64 | Multi-component of signed 64 bit integer |
| sitkVectorFloat32 | Multi-component of 32 bit float |
| sitkVectorFloat64 | Multi-component of 64 bit float |
| sitkLabelUInt8 | RLE label of unsigned 8 bit integers |
| sitkLabelUInt16 | RLE label of unsigned 16 bit integers |
| sitkLabelUInt32 | RLE label of unsigned 32 bit integers |
| sitkLabelUInt64 | RLE label of unsigned 64 bit integers |
There is also sitkUnknown, which is used for undefined or erroneous pixel ID's. It has a value of -1.
The 64-bit integer types are not available on all distributions. When not available the value is sitkUnknown.
import SimpleITK as sitk
# If the environment variable SIMPLE_ITK_MEMORY_CONSTRAINED_ENVIRONMENT is set, this will override the ReadImage
# function so that it also resamples the image to a smaller size (testing environment is memory constrained).
%run setup_for_testing
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
from ipywidgets import interact, fixed
import os
OUTPUT_DIR = "Output"
# Utility method that either downloads data from the Girder repository or
# if already downloaded returns the file name for reading from disk (cached data).
%run update_path_to_download_script
from downloaddata import fetch_data as fdata
Load your first image and display it¶
logo = sitk.ReadImage(fdata("SimpleITK.jpg"))
plt.imshow(sitk.GetArrayViewFromImage(logo))
plt.axis("off");
Fetching SimpleITK.jpg
Image Construction¶
There are a variety of ways to create an image.
The following components are required for a complete definition of an image:
- Pixel type [fixed on creation, no default]: unsigned 32 bit integer, sitkVectorUInt8, etc., see list above.
- Sizes [fixed on creation, no default]: number of pixels/voxels in each dimension. This quantity implicitly defines the image dimension.
- Origin [default is zero]: coordinates of the pixel/voxel with index (0,0,0) in physical units (i.e. mm).
- Spacing [default is one]: Distance between adjacent pixels/voxels in each dimension given in physical units.
- Direction matrix [default is identity]: mapping, rotation, between direction of the pixel/voxel axes and physical directions.
Initial pixel/voxel values are set to zero.
image_3D = sitk.Image(256, 128, 64, sitk.sitkInt16)
image_2D = sitk.Image(64, 64, sitk.sitkFloat32)
image_2D = sitk.Image([32, 32], sitk.sitkUInt32)
image_RGB = sitk.Image([128, 64], sitk.sitkVectorUInt8, 3)
Basic Image Attributes¶
You can change the image origin, spacing and direction using function calls. Making such changes to an image already containing data should be done cautiously.
You can also use the dictionary like bracket operator to make these changes, with the keywords 'origin', 'spacing', 'direction'.
image_3D.SetOrigin((78.0, 76.0, 77.0))
image_3D.SetSpacing([0.5, 0.5, 3.0])
print(f"origin: {image_3D.GetOrigin()}")
print(f"size: {image_3D.GetSize()}")
print(f"spacing: {image_3D.GetSpacing()}")
print(f"direction: {image_3D.GetDirection()}\n")
image_3D["origin"] = (2.0, 4.0, 8.0)
image_3D["spacing"] = [0.25, 0.25, 5.0]
print(f'origin: {image_3D["origin"]}')
print(f"size: {image_3D.GetSize()}")
print(f'spacing: {image_3D["spacing"]}')
print(f'direction: {image_3D["direction"]}')
origin: (78.0, 76.0, 77.0) size: (256, 128, 64) spacing: (0.5, 0.5, 3.0) direction: (1.0, 0.0, 0.0, 0.0, 1.0, 0.0, 0.0, 0.0, 1.0) origin: (2.0, 4.0, 8.0) size: (256, 128, 64) spacing: (0.25, 0.25, 5.0) direction: (1.0, 0.0, 0.0, 0.0, 1.0, 0.0, 0.0, 0.0, 1.0)
Image dimension queries:
print(image_3D.GetDimension())
print(image_3D.GetWidth())
print(image_3D.GetHeight())
print(image_3D.GetDepth())
3 256 128 64
What is the depth of a 2D image?
print(image_2D.GetSize())
print(image_2D.GetDepth())
(32, 32) 0
Pixel/voxel type queries:
print(image_3D.GetPixelIDValue())
print(image_3D.GetPixelIDTypeAsString())
print(image_3D.GetNumberOfComponentsPerPixel())
2 16-bit signed integer 1
What is the dimension and size of a Vector image and its data?
print(image_RGB.GetDimension())
print(image_RGB.GetSize())
print(image_RGB.GetNumberOfComponentsPerPixel())
2 (128, 64) 3
Accessing Pixels and Slicing¶
The Image class's member functions GetPixel and SetPixel provide an ITK-like interface for pixel access.
help(image_3D.GetPixel)
Help on method GetPixel in module SimpleITK.SimpleITK:
GetPixel(*idx) method of SimpleITK.SimpleITK.Image instance
Returns the value of a pixel.
This method takes 2 parameters in 2D: the x and y index,
and 3 parameters in 3D: the x, y and z index.
print(image_3D.GetPixel(0, 0, 0))
image_3D.SetPixel(0, 0, 0, 1)
print(image_3D.GetPixel(0, 0, 0))
# This can also be done using Pythonic notation.
print(image_3D[0, 0, 1])
image_3D[0, 0, 1] = 2
print(image_3D[0, 0, 1])
0 1 0 2
Slicing of SimpleITK images returns a copy of the image data.
This is similar to slicing Python lists and differs from the "view" returned by slicing numpy arrays.
# Brute force sub-sampling
logo_subsampled = logo[::2, ::2]
# Get the sub-image containing the word Simple
simple = logo[0:115, :]
# Get the sub-image containing the word Simple and flip it
simple_flipped = logo[115:0:-1, :]
n = 4
plt.subplot(n, 1, 1)
plt.imshow(sitk.GetArrayViewFromImage(logo))
plt.axis("off")
plt.subplot(n, 1, 2)
plt.imshow(sitk.GetArrayViewFromImage(logo_subsampled))
plt.axis("off")
plt.subplot(n, 1, 3)
plt.imshow(sitk.GetArrayViewFromImage(simple))
plt.axis("off")
plt.subplot(n, 1, 4)
plt.imshow(sitk.GetArrayViewFromImage(simple_flipped))
plt.axis("off");
Draw a square on top of the logo image: After running this cell, uncomment "Version 3" and see its effect.
# Version 0: get the numpy array and assign the value via broadcast - later on you will need to construct
# a new image from the array
logo_pixels = sitk.GetArrayFromImage(logo)
logo_pixels[0:10, 0:10] = [0, 255, 0]
# Version 1: generates an error, cannot assign a multi-channel value to an image, but it does work with one channel
# logo[0:10,0:10] = [255,0,0]
logo_green_channel = sitk.VectorIndexSelectionCast(logo, 1)
logo_green_channel[0:10, 0:10] = 100
sitk.Show(logo_green_channel)
# Version 2: image slicing returns a new image, so all assignments here will not have any effect on the original
# 'logo' image
logo_subimage = logo[0:10, 0:10]
for x in range(0, 10):
for y in range(0, 10):
logo_subimage[x, y] = [255, 0, 0]
# Version 3: modify the original image, iterate and assign a value to each pixel
# for x in range(0,10):
# for y in range(0,10):
# logo[x,y] = [255,0,0]
plt.subplot(2, 1, 1)
plt.imshow(sitk.GetArrayViewFromImage(logo))
plt.axis("off")
plt.subplot(2, 1, 2)
plt.imshow(logo_pixels)
plt.axis("off");
We can also paste one image into the other, either using the PasteImageFilter with its procedural interface or using a more Pythonic approach with image slicing. Note that for these operations SimpleITK treats the images as arrays of pixels and not as spatial objects. In the example below the fact that the images have different spacings is ignored.
logo = sitk.ReadImage(fdata("SimpleITK.jpg"))
sz_x = 10
sz_y = 10
color_channels = [
sitk.Image([sz_x, sz_y], sitk.sitkUInt8),
sitk.Image([sz_x, sz_y], sitk.sitkUInt8) + 255,
sitk.Image([sz_x, sz_y], sitk.sitkUInt8),
]
color_image = sitk.Compose(color_channels)
color_image.SetSpacing([0.5, 0.5])
print(logo.GetSpacing())
print(color_image.GetSpacing())
# Set sub image using the Paste function
logo = sitk.Paste(
destinationImage=logo,
sourceImage=color_image,
sourceSize=color_image.GetSize(),
sourceIndex=[0, 0],
destinationIndex=[0, 0],
)
# Set sub image using slicing.
logo[20 : 20 + sz_x, 0:sz_y] = color_image
sitk.Show(logo)
Fetching SimpleITK.jpg (1.0, 1.0) (0.5, 0.5)
SimpleITK images also support the usage of ellipsis. Below we use both available approaches to obtain a slice.
z_slice = image_3D.GetDepth() // 2
result1 = image_3D[..., z_slice]
result2 = image_3D[:, :, z_slice]
# Check whether the two slices are equivalent, same pixel content and same origin, spacing, direction cosine.
# Uncomment the following line to see what happens if the slices do not have the same origin.
# result1['origin'] = [o+1.0 for o in result1['origin']]
try:
if np.all(sitk.GetArrayFromImage(result1 - result2) == 0):
print("Slices equivalent.")
else:
print("Slices not equivalent (intensity differences).")
except Exception:
print("Slices not equivalent (physical differences).")
Slices equivalent.
Finally, SimpleITK supports masking an image using another image, as shown in the cell below. We are mimicking the deep learning practice of batch normalization.
Comment out the line which addresses the division by zero issue and check the result, hover the cursor over the region which is displayed as black, underlying pixel value isn't zero.
std_positive_threshold = 0.1
image_volume = sitk.ReadImage(fdata("training_001_mr_T1.mha"))
# mimic a batch using slices from a T1 MR volume
batch = [
sitk.Cast(image_volume[:, :, z], sitk.sitkFloat64)
for z in range(image_volume.GetDepth())
]
# perform batch normalization (zero mean, unit standard deviation)
batch_len = len(batch)
batch_mean = sitk.NaryAdd(batch) / batch_len
batch_normalized = [image - batch_mean for image in batch]
batch_sigma = sitk.Sqrt(
sitk.NaryAdd([image**2 for image in batch_normalized]) / batch_len
)
# avoid division by zero using a mask
batch_sigma[batch_sigma < std_positive_threshold] = 1
batch_normalized = [image / batch_sigma for image in batch_normalized]
sitk.Show(sitk.JoinSeries(batch_normalized), "batch normalized images")
Fetching training_001_mr_T1.mha
Conversion between numpy and SimpleITK¶
SimpleITK and numpy indexing access is in opposite order!
SimpleITK: image[x,y,z]
numpy: image_numpy_array[z,y,x]
From SimpleITK to numpy¶
We have two options for converting from SimpleITK to numpy:
- GetArrayFromImage(): returns a copy of the image data. You can then freely modify the data as it has no effect on the original SimpleITK image.
- GetArrayViewFromImage(): returns a view on the image data which is useful for display in a memory efficient manner. You cannot modify the data and the view will be invalid if the original SimpleITK image is deleted.
nda = sitk.GetArrayFromImage(image_3D)
print(image_3D.GetSize())
print(nda.shape)
nda = sitk.GetArrayFromImage(image_RGB)
print(image_RGB.GetSize())
print(nda.shape)
(256, 128, 64) (64, 128, 256) (128, 64) (64, 128, 3)
gabor_image = sitk.GaborSource(size=[64, 64], frequency=0.03)
# Getting a numpy array view on the image data doesn't copy the data
nda_view = sitk.GetArrayViewFromImage(gabor_image)
plt.imshow(nda_view, cmap=plt.cm.Greys_r)
plt.axis("off")
# Trying to assign a value to the array view will throw an exception
nda_view[0, 0] = 255
--------------------------------------------------------------------------- ValueError Traceback (most recent call last) Cell In[17], line 8 5 plt.axis("off") 7 # Trying to assign a value to the array view will throw an exception ----> 8 nda_view[0, 0] = 255 ValueError: assignment destination is read-only
From numpy to SimpleITK¶
Remember to to set the image's origin, spacing, and possibly direction cosine matrix. The default values may not match the physical dimensions of your image.
nda = np.zeros((10, 20, 3))
# if this is supposed to be a 3D gray scale image [x=3, y=20, z=10]
img = sitk.GetImageFromArray(nda)
print(img.GetSize())
# if this is supposed to be a 2D color image [x=20,y=10]
img = sitk.GetImageFromArray(nda, isVector=True)
print(img.GetSize())
(3, 20, 10) (20, 10)
There and back again¶
The following code cell illustrates a situation where your code is a combination of SimpleITK methods and custom Python code which works with intensity values or labels outside of SimpleITK. This is a reasonable approach when you implement an algorithm in Python and don't care about the physical spacing of things (you are actually assuming the volume is isotropic).
def my_algorithm(image_as_numpy_array):
# res is the image result of your algorithm, has the same grid size as the original image
res = image_as_numpy_array
return res
# Starting with SimpleITK
img = sitk.ReadImage(fdata("training_001_mr_T1.mha"))
# Custom Python code working on a numpy array.
npa_res = my_algorithm(sitk.GetArrayFromImage(img))
# Converting back to SimpleITK (assumes we didn't move the image in space as we copy the information from the original)
res_img = sitk.GetImageFromArray(npa_res)
res_img.CopyInformation(img)
# Continuing to work with SimpleITK images
res_img - img
Fetching training_001_mr_T1.mha
<SimpleITK.SimpleITK.Image; proxy of <Swig Object of type 'itk::simple::Image *' at 0x10b20d800> >
Image operations¶
SimpleITK supports basic arithmetic operations between images, taking into account their physical space.
Repeatedly run this cell. Fix the error (comment out the SetDirection, then SetSpacing). Why doesn't the SetOrigin line cause a problem? How close do two physical attributes need to be in order to be considered equivalent?
img1 = sitk.Image(24, 24, sitk.sitkUInt8)
img1[0, 0] = 0
img2 = sitk.Image(img1.GetSize(), sitk.sitkUInt8)
img2.SetDirection([0, 1, 0.5, 0.5])
img2.SetSpacing([0.5, 0.8])
img2.SetOrigin([0.000001, 0.000001])
img2[0, 0] = 255
img3 = img1 + img2
print(img3[0, 0])
--------------------------------------------------------------------------- RuntimeError Traceback (most recent call last) Cell In[20], line 10 7 img2.SetOrigin([0.000001, 0.000001]) 8 img2[0, 0] = 255 ---> 10 img3 = img1 + img2 11 print(img3[0, 0]) File ~/toolkits/anaconda3/envs/sitkMaster3.11/lib/python3.11/site-packages/SimpleITK/SimpleITK.py:3968, in Image.__add__(self, other) 3966 def __add__( self, other ): 3967 if isinstance( other, Image ): -> 3968 return Add( self, other ) 3969 try: 3970 return Add( self, float(other) ) File ~/toolkits/anaconda3/envs/sitkMaster3.11/lib/python3.11/site-packages/SimpleITK/SimpleITK.py:11631, in Add(*args) 11623 def Add(*args): 11624 r""" 11625 Add(Image image1, Image image2) -> Image 11626 Add(Image image1, double constant) -> Image (...) 11629 11630 """ > 11631 return _SimpleITK.Add(*args) RuntimeError: Exception thrown in SimpleITK Add: /Users/runner/work/SimpleITK/SimpleITK/bld/ITK-prefix/include/ITK-5.4/itkImageToImageFilter.hxx:217: ITK ERROR: AddImageFilter(0x7fa1b7806520): Inputs do not occupy the same physical space! InputImage Spacing: [1.0000000e+00, 1.0000000e+00], InputImage_1 Spacing: [5.0000000e-01, 8.0000000e-01] Tolerance: 1.0000000e-06 InputImage Direction: 1.0000000e+00 0.0000000e+00 0.0000000e+00 1.0000000e+00 , InputImage_1 Direction: 0.0000000e+00 1.0000000e+00 5.0000000e-01 5.0000000e-01 Tolerance: 1.0000000e-06
Reading and Writing¶
SimpleITK can read and write images stored in a single file, or a set of files (e.g. DICOM series).
Images stored in the DICOM format have a meta-data dictionary associated with them, which is populated with the DICOM tags. When a DICOM series is read as a single image, the meta-data information is not available since DICOM tags are specific to each file. If you need the meta-data, you have three options:
Using the object oriented interface's ImageSeriesReader class, configure it to load the tags using the
MetaDataDictionaryArrayUpdateOnmethod and possibly theLoadPrivateTagsOnmethod if you need the private tags. Once the series is read you can access the meta-data from the series reader using theGetMetaDataKeys,HasMetaDataKey, andGetMetaData.Using the object oriented interface's ImageFileReader, set a specific slice's file name and only read it's meta-data using the
ReadImageInformationmethod which only reads the meta-data but not the bulk pixel information. Once the meta-data is read you can access it from the file reader using theGetMetaDataKeys,HasMetaDataKey, andGetMetaData.Using the object oriented interface's ImageFileReader, set a specific slice's file name and read it. Or using the procedural interface's, ReadImage function, read a specific file. You can then access the meta-data directly from the Image using the
GetMetaDataKeys,HasMetaDataKey, andGetMetaData.
In the following cell, we read an image in JPEG format, and write it as PNG and BMP. File formats are deduced from the file extension. Appropriate pixel type is also set - you can override this and force a pixel type of your choice.
Note: When reading an image series, via the ImageSeriesReader or via the procedural ReadImage interface, the images in the list are assumed to be ordered correctly (GetGDCMSeriesFileNames ensures this for DICOM). If the order is incorrect, the image will be read, but its spacing and possibly the direction cosine matrix will be incorrect.
img = sitk.ReadImage(fdata("SimpleITK.jpg"))
print(img.GetPixelIDTypeAsString())
# write as PNG and BMP
sitk.WriteImage(img, os.path.join(OUTPUT_DIR, "SimpleITK.png"))
sitk.WriteImage(img, os.path.join(OUTPUT_DIR, "SimpleITK.bmp"))
Fetching SimpleITK.jpg vector of 8-bit unsigned integer
Read an image in JPEG format and cast the pixel type according to user selection.
# Several pixel types, some make sense in this case (vector types) and some are just show
# that the user's choice will force the pixel type even when it doesn't make sense
# (e.g. sitkVectorUInt16 or sitkUInt8).
pixel_types = {
"sitkUInt8": sitk.sitkUInt8,
"sitkUInt16": sitk.sitkUInt16,
"sitkFloat64": sitk.sitkFloat64,
"sitkVectorUInt8": sitk.sitkVectorUInt8,
"sitkVectorUInt16": sitk.sitkVectorUInt16,
"sitkVectorFloat64": sitk.sitkVectorFloat64,
}
def pixel_type_dropdown_callback(pixel_type, pixel_types_dict):
# specify the file location and the pixel type we want
img = sitk.ReadImage(fdata("SimpleITK.jpg"), pixel_types_dict[pixel_type])
print(img.GetPixelIDTypeAsString())
print(img[0, 0])
plt.imshow(sitk.GetArrayViewFromImage(img))
plt.axis("off")
interact(
pixel_type_dropdown_callback,
pixel_type=list(pixel_types.keys()),
pixel_types_dict=fixed(pixel_types),
);
Read a DICOM series and write it as a single mha file
data_directory = os.path.dirname(fdata("CIRS057A_MR_CT_DICOM/readme.txt"))
series_ID = "1.2.840.113619.2.290.3.3233817346.783.1399004564.515"
# Get the list of files belonging to a specific series ID.
reader = sitk.ImageSeriesReader()
# Use the functional interface to read the image series.
original_image = sitk.ReadImage(
reader.GetGDCMSeriesFileNames(data_directory, series_ID)
)
# Write the image.
output_file_name_3D = os.path.join(OUTPUT_DIR, "3DImage.mha")
sitk.WriteImage(original_image, output_file_name_3D)
# Read it back again.
written_image = sitk.ReadImage(output_file_name_3D)
# Check that the original and written image are the same.
statistics_image_filter = sitk.StatisticsImageFilter()
statistics_image_filter.Execute(original_image - written_image)
# Check that the original and written files are the same
print(
f"Max, Min differences are : {statistics_image_filter.GetMaximum()}, {statistics_image_filter.GetMinimum()}"
)
Fetching CIRS057A_MR_CT_DICOM/readme.txt
Max, Min differences are : 0.0, 0.0
Write an image series as JPEG. The WriteImage function receives a volume and a list of images names and writes the volume according to the z axis. For a displayable result we need to rescale the image intensities (default is [0,255]) since the JPEG format requires a cast to the UInt8 pixel type.
sitk.WriteImage(
sitk.Cast(sitk.RescaleIntensity(written_image), sitk.sitkUInt8),
[
os.path.join(OUTPUT_DIR, f"slice{i:03d}.jpg")
for i in range(written_image.GetSize()[2])
],
)
Select a specific DICOM series from a directory and only then load user selection.
data_directory = os.path.dirname(fdata("CIRS057A_MR_CT_DICOM/readme.txt"))
# Global variable 'selected_series' is updated by the interact function
selected_series = ""
file_reader = sitk.ImageFileReader()
def DICOM_series_dropdown_callback(series_to_load, series_dictionary):
global selected_series
# Print some information about the series from the meta-data dictionary
# DICOM standard part 6, Data Dictionary: http://medical.nema.org/medical/dicom/current/output/pdf/part06.pdf
file_reader.SetFileName(series_dictionary[series_to_load][0])
file_reader.ReadImageInformation()
tags_to_print = {
"0010|0010": "Patient name: ",
"0008|0060": "Modality: ",
"0008|0021": "Series date: ",
"0008|0080": "Institution name: ",
"0008|1050": "Performing physician's name: ",
}
for tag in tags_to_print:
try:
print(tags_to_print[tag] + file_reader.GetMetaData(tag))
except: # Ignore if the tag isn't in the dictionary
pass
selected_series = series_to_load
# Directory contains multiple DICOM studies/series, store
# in dictionary with key being the series ID
reader = sitk.ImageSeriesReader()
series_file_names = {}
series_IDs = reader.GetGDCMSeriesIDs(data_directory)
# Check that we have at least one series
if series_IDs:
for series in series_IDs:
series_file_names[series] = reader.GetGDCMSeriesFileNames(
data_directory, series
)
interact(
DICOM_series_dropdown_callback,
series_to_load=list(series_IDs),
series_dictionary=fixed(series_file_names),
)
else:
print("Data directory does not contain any DICOM series.")
Fetching CIRS057A_MR_CT_DICOM/readme.txt
reader.SetFileNames(series_file_names[selected_series])
img = reader.Execute()
# Display the image slice from the middle of the stack, z axis
z = int(img.GetDepth() / 2)
plt.imshow(sitk.GetArrayViewFromImage(img)[z, :, :], cmap=plt.cm.Greys_r)
plt.axis("off");
DICOM photometric interpretation¶
Generally speaking, SimpleITK represents color images as multi-channel images independent of a color space. It is up to you to interpret the channels correctly based on additional color space knowledge prior to using them for display or any other purpose.
The following cells illustrate reading and interpretation of interesting images in DICOM format. The first is a photograph of an X-ray on a light box (yes, there are some strange things in the wild). The second is a digital X-ray. While both of these are chest X-rays they differ in image modality (0008|0060) and in Photometric Interpretation (0028|0004), color space in DICOM speak.
Things to note:
- When using SimpleITK to read a color DICOM image, the channel values will be transformed to the RGB color space.
- When using SimpleITK to read a scalar image, it is assumed that the lowest intensity value is black and highest white. If the photometric interpretation tag is MONOCHROME2 (lowest value displayed as black) nothing is done. If it is MONOCHROME1 (lowest value displayed as white), the pixel values are inverted.
xrays = [sitk.ReadImage(fdata("photo.dcm")), sitk.ReadImage(fdata("cxr.dcm"))]
# We can access the image's metadata via the GetMetaData method or
# via the bracket operator, the latter is more concise.
for img in xrays:
print(f'Image Modality: {img.GetMetaData("0008|0060")}')
print(f"Number of channels: {img.GetNumberOfComponentsPerPixel()}")
print(f'Photomertic Interpretation: {img["0028|0004"]}')
# Display the image using Fiji which expects the channels to be in the RGB color space
sitk.Show(img)
Fetching photo.dcm Fetching cxr.dcm
Image Modality: XC Number of channels: 3 Photomertic Interpretation: YBR_FULL_422 Image Modality: CR Number of channels: 1 Photomertic Interpretation: MONOCHROME1
WARNING: In /Users/runner/work/SimpleITK/SimpleITK/bld/ITK/Modules/IO/GDCM/src/itkGDCMImageIO.cxx, line 361 GDCMImageIO (0x7fa1a730bdb0): Converting from MONOCHROME1 to MONOCHROME2 may impact the meaning of DICOM attributes related to pixel values. WARNING: In /Users/runner/work/SimpleITK/SimpleITK/bld/ITK/Modules/IO/Meta/src/itkMetaImageIO.cxx, line 660 MetaImageIO (0x7fa2500afc00): Unsupported or empty metaData item 0008|0050 of type NSt3__112basic_stringIcNS_11char_traitsIcEENS_9allocatorIcEEEEfound, won't be written to image file WARNING: In /Users/runner/work/SimpleITK/SimpleITK/bld/ITK/Modules/IO/Meta/src/itkMetaImageIO.cxx, line 660 MetaImageIO (0x7fa2500afc00): Unsupported or empty metaData item 0018|0010 of type NSt3__112basic_stringIcNS_11char_traitsIcEENS_9allocatorIcEEEEfound, won't be written to image file WARNING: In /Users/runner/work/SimpleITK/SimpleITK/bld/ITK/Modules/IO/Meta/src/itkMetaImageIO.cxx, line 660 MetaImageIO (0x7fa2500afc00): Unsupported or empty metaData item 0018|5101 of type NSt3__112basic_stringIcNS_11char_traitsIcEENS_9allocatorIcEEEEfound, won't be written to image file WARNING: In /Users/runner/work/SimpleITK/SimpleITK/bld/ITK/Modules/IO/Meta/src/itkMetaImageIO.cxx, line 660 MetaImageIO (0x7fa2500afc00): Unsupported or empty metaData item 0020|0010 of type NSt3__112basic_stringIcNS_11char_traitsIcEENS_9allocatorIcEEEEfound, won't be written to image file WARNING: In /Users/runner/work/SimpleITK/SimpleITK/bld/ITK/Modules/IO/Meta/src/itkMetaImageIO.cxx, line 660 MetaImageIO (0x7fa2500afc00): Unsupported or empty metaData item 0020|0020 of type NSt3__112basic_stringIcNS_11char_traitsIcEENS_9allocatorIcEEEEfound, won't be written to image file WARNING: In /Users/runner/work/SimpleITK/SimpleITK/bld/ITK/Modules/IO/Meta/src/itkMetaImageIO.cxx, line 660 MetaImageIO (0x7fa2500afc00): Unsupported or empty metaData item 0020|0060 of type NSt3__112basic_stringIcNS_11char_traitsIcEENS_9allocatorIcEEEEfound, won't be written to image file WARNING: In /Users/runner/work/SimpleITK/SimpleITK/bld/ITK/Modules/IO/Meta/src/itkMetaImageIO.cxx, line 660 MetaImageIO (0x7fa2500afc00): Unsupported or empty metaData item 0032|4000 of type NSt3__112basic_stringIcNS_11char_traitsIcEENS_9allocatorIcEEEEfound, won't be written to image file
The first, is a color sRGB image while an x-ray should be a single channel gray scale image. We will convert sRGB to gray scale.
def srgb2gray(image):
# Convert sRGB image to gray scale and rescale results to [0,255]
channels = [
sitk.VectorIndexSelectionCast(image, i, sitk.sitkFloat32)
for i in range(image.GetNumberOfComponentsPerPixel())
]
# linear mapping
I = 1 / 255.0 * (0.2126 * channels[0] + 0.7152 * channels[1] + 0.0722 * channels[2])
# nonlinear gamma correction
I = (
I * sitk.Cast(I <= 0.0031308, sitk.sitkFloat32) * 12.92
+ I ** (1 / 2.4) * sitk.Cast(I > 0.0031308, sitk.sitkFloat32) * 1.055
- 0.055
)
return sitk.Cast(sitk.RescaleIntensity(I), sitk.sitkUInt8)
sitk.Show(srgb2gray(xrays[0]))
Finer control¶
The ImageFileReader's interface provides finer control for reading, allowing us to require the use of a specific IO and allowing us to stream parts of an image to memory without reading the whole image (supported by a subset of the ImageIO components).
Selecting a Specific Image IO¶
SimpleITK relies on the registered ImageIOs to indicate whether they can read a file and then perform the reading. This is done automatically, going over the set of ImageIOs and inquiring whether they can read the given file. The first one that can is selected. If multiple ImageIOs can read a specific format, we do not know which one was used for the task (e.g. TIFFImageIO and LSMImageIO, which is derived from it, can both read tif files). In some cases you may want to use a specific IO, possibly one that reads the file faster, or supports a more complete feature set associated with the file format.
The next cell shows how to find out which ImageIOs are registered and specify the one we want.
file_reader = sitk.ImageFileReader()
# Get a tuple listing all registered ImageIOs
image_ios_tuple = file_reader.GetRegisteredImageIOs()
print("The supported image IOs are: " + str(image_ios_tuple))
# Optionally, just print the reader and see which ImageIOs are registered
print("\n", file_reader)
# Specify the JPEGImageIO and read file
file_reader.SetImageIO("JPEGImageIO")
file_reader.SetFileName(fdata("SimpleITK.jpg"))
logo = file_reader.Execute()
# Unfortunately, now reading a non JPEG image will fail
try:
file_reader.SetFileName(fdata("cthead1.png"))
ct_head = file_reader.Execute()
except RuntimeError:
print("Got a RuntimeError exception.")
# We can reset the file reader to its default behaviour so that it automatically
# selects the ImageIO
file_reader.SetImageIO("")
ct_head = file_reader.Execute()
The supported image IOs are: ('BMPImageIO', 'BioRadImageIO', 'Bruker2dseqImageIO', 'GDCMImageIO', 'GE4ImageIO', 'GE5ImageIO', 'GiplImageIO', 'HDF5ImageIO', 'JPEGImageIO', 'JPEG2000ImageIO', 'LSMImageIO', 'MINCImageIO', 'MRCImageIO', 'MetaImageIO', 'NiftiImageIO', 'NrrdImageIO', 'PNGImageIO', 'StimulateImageIO', 'TIFFImageIO', 'VTKImageIO')
itk::simple::ImageFileReader
FileName: ""
ExtractSize: [ ]
ExtractIndex: [ ]
Image Information:
PixelType: Unknown pixel id
Dimension: 0
NumberOfComponents: 0
Direction: [ ]
Origin: [ ]
Spacing: [ ]
Size: [ ]
OutputPixelType: Unknown pixel id
LoadPrivateTags: 0
ImageIOName:
Registered ImageIO:
BMPImageIO ( *.bmp, *.BMP, )
BioRadImageIO ( *.PIC, *.pic, )
Bruker2dseqImageIO
GDCMImageIO ( *.dcm, *.DCM, *.dicom, *.DICOM, )
GE4ImageIO
GE5ImageIO
GiplImageIO
HDF5ImageIO ( *.hdf, *.h4, *.hdf4, *.h5, *.hdf5, *.he4, *.he5, *.hd5, )
JPEGImageIO ( *.jpg, *.JPG, *.jpeg, *.JPEG, )
JPEG2000ImageIO ( *.j2k, *.jp2, *.jpt, )
LSMImageIO ( *.lsm, *.LSM, )
MINCImageIO ( *.mnc, *.MNC, *.mnc2, *.MNC2, )
MRCImageIO ( *.mrc, *.rec, )
MetaImageIO ( *.mha, *.mhd, )
NiftiImageIO ( *.nia, *.nii, *.nii.gz, *.hdr, *.img, *.img.gz, )
NrrdImageIO ( *.nrrd, *.nhdr, )
PNGImageIO ( *.png, *.PNG, )
StimulateImageIO ( *.spr, )
TIFFImageIO ( *.tif, *.TIF, *.tiff, *.TIFF, )
VTKImageIO ( *.vtk, )
Debug: 0
NumberOfThreads: 16
NumberOfWorkUnits: 0
Commands: (none)
ProgressMeasurement: 0
ActiveProcess: (none)
Fetching SimpleITK.jpg
Fetching cthead1.png
Got a RuntimeError exception.
Not a JPEG file: starts with 0x89 0x50 libpng warning: sCAL: invalid unit libpng warning: sCAL: invalid unit libpng warning: sCAL: invalid unit
Streaming Image IO¶
Some of the ImageIOs supported in SimpleITK allow you to stream in sub-regions of an image without the need to read the whole image into memory. This is very useful when you are memory constrained (either your images are large or your memory is limited).
The ImageIOs that support streaming include HDF5ImageIO, VTKImageIO, NiftiImageIO, MetaImageIO...
The next cell shows how to read in a sub/cropped image from a larger image. We read the central 1/3 portion of the image [1/3,2/3] of the original image.
file_reader = sitk.ImageFileReader()
file_reader.SetFileName(fdata("vm_head_rgb.mha"))
file_reader.ReadImageInformation()
image_size = file_reader.GetSize()
start_index, extract_size = zip(
*[(int(1.0 / 3.0 * sz), int(1.0 / 3.0 * sz)) for sz in file_reader.GetSize()]
)
file_reader.SetExtractIndex(start_index)
file_reader.SetExtractSize(extract_size)
sitk.Show(file_reader.Execute())
Fetching vm_head_rgb.mha
The next cells show how to subtract two large images from each other with a smaller memory footprint than the direct approach, though the code is much more complex and slower than the direct approach:
sitk.ReadImage(image1_file_name) - sitk.ReadImage(image2_file_name)
Note: The code assume that the two images occupy the same spatial region (origin, spacing, direction cosine matrix).
def streaming_subtract(image1_file_name, image2_file_name, parts):
"""
Subtract image1 from image2 using 'parts' number of sub-regions.
"""
file_reader = sitk.ImageFileReader()
file_reader.SetFileName(image1_file_name)
file_reader.ReadImageInformation()
image_size = file_reader.GetSize()
# Create the result image, initially empty
result_img = sitk.Image(
file_reader.GetSize(),
file_reader.GetPixelID(),
file_reader.GetNumberOfComponents(),
)
result_img.SetSpacing(file_reader.GetSpacing())
result_img.SetOrigin(file_reader.GetOrigin())
result_img.SetDirection(file_reader.GetDirection())
extract_size = list(file_reader.GetSize())
extract_size[-1] = extract_size[-1] // parts
current_index = [0] * file_reader.GetDimension()
for i in range(parts):
if i == (
parts - 1
): # last region may be smaller than the standard extract region
extract_size[-1] = image_size[-1] - current_index[-1]
file_reader.SetFileName(image1_file_name)
file_reader.SetExtractIndex(current_index)
file_reader.SetExtractSize(extract_size)
sub_image1 = file_reader.Execute()
file_reader.SetFileName(image2_file_name)
file_reader.SetExtractIndex(current_index)
file_reader.SetExtractSize(extract_size)
sub_image2 = file_reader.Execute()
# Paste the result of subtracting the two subregions into their location in the result_img
result_img = sitk.Paste(
result_img,
sub_image1 - sub_image2,
extract_size,
[0] * file_reader.GetDimension(),
current_index,
)
current_index[-1] += extract_size[-1]
return result_img
# If you have the patience and RAM you can try this with the vm_head_rgb.mha image.
image1_file_name = fdata("fib_sem_bacillus_subtilis.mha")
image2_file_name = fdata("fib_sem_bacillus_subtilis.mha")
Fetching fib_sem_bacillus_subtilis.mha Fetching fib_sem_bacillus_subtilis.mha
A simple way of seeing your system's memory usage is to open the appropriate monitoring program: (Windows) Resource Monitor; (Linux) top; (OS X) Activity Monitor. This will give you a rough idea of the memory used by the streaming vs. non streaming approaches.
result_img = streaming_subtract(image1_file_name, image2_file_name, parts=5)
del result_img
result_img = sitk.ReadImage(image1_file_name) - sitk.ReadImage(image2_file_name)
del result_img
Reading from memory¶
ITK and consequentially SimpleITK support file based IO. Therefore, to read file data that is in memory we need a workaround. In Python this is relatively straightforward using the tempfile module and function decorators as shown below.
Note that the same decorator can be used to read a Transformation from memory (decorate the ReadTransform function).
When speed is of the essence, we can override the default temp directory and point it to a RAM disk based file system, speeding up IO. On Linux systems /dev/shm provides a RAM based file system and we can point the default tempdir to that directory (be careful as the memory size is more restricted).
Technical Note: Windows has a bug with temporary files. On windows, when creating NamedTemporaryFile, if delete=True the file is kept open and cannot be read from. We set the delete flag to False on windows, delete=os.name != "nt", and True on all other operating systems, circumventing the issue (there needs to be a cleanup step in the end, which isn't implemented here).
import requests
import tempfile
import os
# override the default temp directory with a user specified one
# See details: https://docs.python.org/3/library/tempfile.html#tempfile.gettempdir
#
# os.environ["TMPDIR"] = "path/to/ram/disk/dir"
def bytes_decorator(file_extension):
"""
A decorator which takes a SimpleITK function which expects a file name and replaces it with one that expects
a binary blob. The decorator writes a temporary file with the given suffix and the wrapped function is invoked
with the file. Using the suffix is critical, because the SimpleITK introspection mechanism checks which
IO component can read a file. Some IO components (e.g MetaImageIO) rely on the file extension.
"""
def decorator(func):
def wrapper(*args, **kwargs):
with tempfile.NamedTemporaryFile(
mode="wb", suffix=file_extension, delete=os.name != "nt"
) as fp:
fp.write(args[0])
fp.flush()
args_list = list(args)
args_list[0] = fp.name
return func(*args_list, **kwargs)
return wrapper
return decorator
sitk.ReadMemoryJPG = bytes_decorator(".jpg")(sitk.ReadImage)
# Get binary data from the SimpleITK website
response = requests.get(
"https://simpleitk.org/images/SimpleITK-Icons/s-name-full-white.jpg"
)
if response.ok:
sitk.Show(sitk.ReadMemoryJPG(response.content))
The decorator above uses default settings for the underlying ImageFileReader and creates a reader every time the function is invoked. In some cases this is not sufficiently flexible. The code below illustrates an alternative solution which provides the additional flexibility.
def read_file_from_memory(file_reader, memory_file_blob):
"""
Read the binary data as a file. Binary data is written to disk using the tempfile
mechanism and the resulting file is read using the pre-configured FileReader. It is
critical to set the FileReader's specific IO (SetImageIO) to match the type of the file.
Because the file is written without an extension, SimpleITK cannot use its introspection
mechanism to automatically identify which IO can read the file because some IOs (e.g. MetaImageIO)
require a specific file extension, otherwise they claim that they cannot read the file.
Useful for reading images.
"""
with tempfile.NamedTemporaryFile(mode="wb", delete=os.name != "nt") as fp:
fp.write(memory_file_blob)
fp.flush()
file_reader.SetFileName(fp.name)
return file_reader.Execute()
image_file_reader = sitk.ImageFileReader()
image_file_reader.SetImageIO("GDCMImageIO")
image_file_reader.LoadPrivateTagsOn()
sitk.ReadMemoryDCM = bytes_decorator(".dcm")(sitk.ReadImage)
# read file into memory
with open(fdata("cxr.dcm"), "rb") as fp:
binary_data = fp.read()
# read from memory using the two options
image_opt1 = sitk.ReadMemoryDCM(binary_data)
image_opt2 = read_file_from_memory(image_file_reader, binary_data)
# The group number (first part of the group|element) of a private
# tag is an odd number. Print the dictionaries for the two options and
# see that the second option includes private tags while the first
# does not.
print(image_opt1.GetMetaDataKeys())
print(image_opt2.GetMetaDataKeys())
Fetching cxr.dcm
WARNING: In /Users/runner/work/SimpleITK/SimpleITK/bld/ITK/Modules/IO/GDCM/src/itkGDCMImageIO.cxx, line 361 GDCMImageIO (0x7fa136c14f90): Converting from MONOCHROME1 to MONOCHROME2 may impact the meaning of DICOM attributes related to pixel values.
('0008|0005', '0008|0008', '0008|0016', '0008|0018', '0008|0020', '0008|0021', '0008|0022', '0008|0023', '0008|0030', '0008|0031', '0008|0032', '0008|0033', '0008|0050', '0008|0060', '0008|1010', '0018|0010', '0018|0015', '0018|1004', '0018|1164', '0018|1400', '0018|1401', '0018|1508', '0018|5101', '0018|6000', '0020|000d', '0020|000e', '0020|0010', '0020|0011', '0020|0012', '0020|0013', '0020|0020', '0020|0060', '0028|0002', '0028|0004', '0028|0010', '0028|0011', '0028|0030', '0028|0100', '0028|0101', '0028|0102', '0028|0103', '0028|1050', '0028|1051', '0028|2110', '0032|1033', '0032|4000', '2010|0010', '2010|0030', '2010|0040', '2010|0100', '2010|0140', '2050|0020', 'ITK_original_direction', 'ITK_original_spacing')
('0008|0005', '0008|0008', '0008|0016', '0008|0018', '0008|0020', '0008|0021', '0008|0022', '0008|0023', '0008|0030', '0008|0031', '0008|0032', '0008|0033', '0008|0050', '0008|0060', '0008|1010', '0009|0010', '0009|1004', '0009|1005', '0009|1006', '0009|1008', '0009|1009', '0009|100c', '0009|1010', '0009|1080', '0009|1090', '0009|1092', '0009|10f0', '0009|10f1', '0018|0010', '0018|0015', '0018|1004', '0018|1164', '0018|1400', '0018|1401', '0018|1508', '0018|5101', '0018|6000', '0019|0010', '0019|1015', '0019|1032', '0019|1040', '0019|1050', '0019|1060', '0019|1080', '0019|1081', '0019|1090', '0019|1091', '0020|000d', '0020|000e', '0020|0010', '0020|0011', '0020|0012', '0020|0013', '0020|0020', '0020|0060', '0021|0010', '0021|1010', '0021|1030', '0021|1040', '0021|1050', '0025|0010', '0025|1010', '0025|1011', '0025|1012', '0028|0002', '0028|0004', '0028|0010', '0028|0011', '0028|0030', '0028|0100', '0028|0101', '0028|0102', '0028|0103', '0028|1050', '0028|1051', '0028|2110', '0029|0010', '0029|1020', '0029|1030', '0029|1034', '0029|1044', '0029|1050', '0032|1033', '0032|4000', '2010|0010', '2010|0030', '2010|0040', '2010|0100', '2010|0140', '2050|0020', '50f1|0010', '50f1|100a', '50f1|1010', '50f1|1020', 'ITK_original_direction', 'ITK_original_spacing')
WARNING: In /Users/runner/work/SimpleITK/SimpleITK/bld/ITK/Modules/IO/GDCM/src/itkGDCMImageIO.cxx, line 361 GDCMImageIO (0x7fa1e6fe8710): Converting from MONOCHROME1 to MONOCHROME2 may impact the meaning of DICOM attributes related to pixel values.